Tuberculosis surveillance for a world health crisis
Tuberculosis (TB) surveillance is a vital step toward eradicating a disease that is the second leading infectious disease killer worldwide, behind COVID-19.1 Caused by bacteria in the Mycobacterium tuberculosis complex (MTBC), which includes Mycobacterium tuberculosis (Mtb), TB is a treatable disease but multidrug-resistant TB (MDR-TB) remains a public health crisis and a health security threat.1 Genomic-based TB surveillance can support public health officials in:
- Detecting Mtb and non-TB species
- Characterizing anti-TB drug resistance
- Tracking the path of transmission
- Monitoring the evolution of the mycobacterium and novel mutations that are resistant to existing and new forms of treatment
The discriminatory power of next-generation sequencing enables outbreaks to be addressed with greater speed and confidence.
Genomic analysis for detecting drug-resistant tuberculosis
Drug-resistant TB (DR-TB) is an MTBC that is resistant to one or more anti-TB drugs and represents a major challenge in controlling the pathogen’s diagnosis, treatment, and eradication.2 Detecting and characterizing these strains are therefore a vital part of disease surveillance.
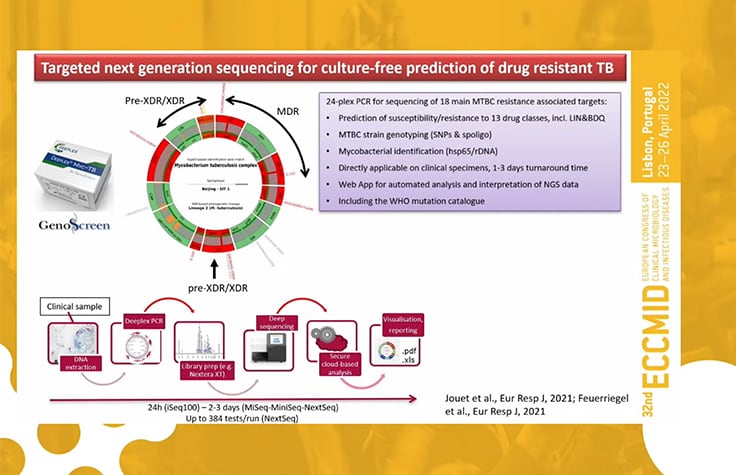
Next-generation sequencing (NGS)-based solutions, such as targeted NGS (tNGS) and whole-genome sequencing (WGS), offer key advantages in detecting DR-TB and are detailed below.
Targeted next-generation sequencing advantages
Sample preparation
- Start directly from specimens, no need for timely cultures
Detection capabilities
- Genotyping and spoligotyping of Mtb
- Target drug resistance markers for a wide range of anti-TB drug resistance detection
- Detection of heteroresistant subpopulations
Focused detection
- Detection of Mtb and non-TB mycobacterium strains
- Accurate characterization of nucleotide-level genetic polymorphisms
Whole-genome sequencing advantages
Sample preparation
- Start from cultured isolates
Detection capabilities
- Full genome sequence data for variant detection and resistant mechanisms, regardless of prior knowledge
Focused detection
- Unbiased characterization of entire genomes for high resolution epidemiological investigations
- Full genome available for any analysis needs
Tuberculosis Resources
Tuberculosis infographic
This comprehensive infographic displays information about TB drugs, treatment, surveillance methods, and more facts and figures in a visual format.

Direct bacterial colony sequencing
Eliminate the need for separate DNA isolation and high-accuracy quantitation steps with the streamlined, cost-effective Flex Direct Colony Method.
Featured Product
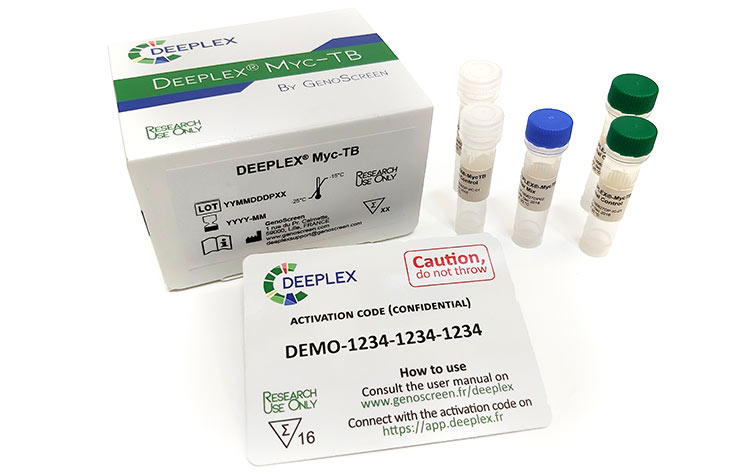
Illumina and Genoscreen Deeplex Myc-TB Combo Kit
A comprehensive, culture-free solution to characterize TB drug resistance with fast results in less than 48 hours.
Learn MoreRelated Products

MiSeq System and Reagents
The MiSeq benchtop sequencer enables targeted and microbial genome applications, with high-quality sequencing, simple data analysis, and cloud storage.

MiniSeq System
Explore how the MiniSeq System can harness the power of NGS in an accessible sequencing format perfect for your benchtop. Streamlined and cost-efficient for rapid sequencing of DNA and RNA.

Illumina DNA Prep
The Illumina DNA Prep is a fast, user-friendly solution for a wide range of applications from human whole-genome sequencing to amplicons, plasmids, and microbial species.

iSeq System
The iSeq 100 Sequencing System makes next-generation sequencing easier and more affordable than ever. Designed for simplicity, it allows labs of all sizes to sequence DNA and RNA at the push of a button.
Interested in learning more about TB surveillance products?
Fill out the form below and we'll be in touch.Moving next-generation sequencing to clinical microbiology laboratories
The ECCMID 2022 Symposium is now available. Learn about metagenomics, genomic surveillance, microbiomes, and next-generation sequencing for tuberculosis.
Watch videoAdditional Resources
Using genomics for tuberculosis surveillance
In this featured article, read how Dr. Camilla Rodrigues is using NGS to fight MDR-TB at the P.D. Hinduja Hospital and Medical Research Centre in Mumbai, India.
Seeking the source of bacterial drug resistance
Read the interview with Dr. Supply, who discusses the growing problem of MDR-TB, the values that Illumina next-generation sequencing (NGS) systems bring to their detection, and more.
Shaheed Vally Omar iCommunity article
Read what the scientific lead at South Africa’s Centre for Tuberculosis in Johannesburg believes when it comes to detecting and tracking MDR-TB using next-generation sequencing.
NGS in clinical microbiology research
In this webinar, Dr. Arryn Craney, discusses integration options for NGS in microbiology clinical research and her recent hands-on experience with the Respiratory Pathogen ID/AMR Enrichment Panel (RPIP) Kit. In addition, this webinar explores future possibilities for advanced diagnostics.
World Tuberculosis Day: March 24
World TB Day commemorates the day Dr. Robert Koch announced the bacterium responsible for TB in 1882. This day raises public awareness about the devastating impact TB continues to have and the challenges that lay ahead as we drive toward control and prevention.
References
- who.int/news-room/fact-sheets/detail/tuberculosisWHO.com
- Castro RAD, Borrell S, Gagneux S. The within-host evolution of antimicrobial resistance in Mycobacterium tuberculosis. FEMS Microbiol Rev. 2021 Aug 17;45(4) doi: 10.1093/femsre/fuaa071